Ming C. Hammond
 ORGANIC & BIOLOGICAL CHEMISTRY
ORGANIC & BIOLOGICAL CHEMISTRY
Professor of Chemistry
Executive Committee, Henry Eyring Center for Cell and Genome Science
Co-Director, U of U Beckman Scholar Program
Co-Director, Utah Chemistry REU Program
B. S. California Institute of Technology, 2000
Ph. D. University of California, Berkeley, 2005
Burroughs Wellcome Fund Postdoctoral Fellow, Yale University, 2005-2009
Phone: 801-213-0892
Office: 224B Crocker Science Center
Email: mingch@chem.utah.edu
Research Group
Activities & Awards
- 2016 Women in Science Award, Chau Hoi Shuen Foundation
- 2011-2016 NIH Director’s New Innovator Award
- 2011 Regents’ Junior Faculty Fellowship, UC Berkeley
- 2010 Presidential Chair Fellow, UC Berkeley
- 2010-2013 Chevron Chair of Chemistry, UC Berkeley
- 2010 Thieme Chemistry Journal Award
- 2008-2015 Career Award at the Scientific Interface, Burroughs Wellcome Fund
- 2000-2005 HHMI Predoctoral Fellowship
- 2000 NSF Graduate Research Fellowship (declined)
- 2000 Richard P. Schuster Memorial Prize in Chemistry, Caltech
- 1999-2000 Carnation Merit Award, Caltech
- 1999 Arie J. Haagen-Smith Memorial Award in Chemistry, Caltech
- 1998-1999 Beckman Scholar Fellowship
- 1998-1999 Caltech Prize Award
Research Interests
Illuminating the Single-Cell Biology and Function of Chemical Signals
The Hammond lab has a dual focus on engineering nucleic acids as programmable tools
for molecular imaging and gene control, and on understanding the chemistry and biology
of nucleotide-based signaling molecules in bacteria and mammalian cells.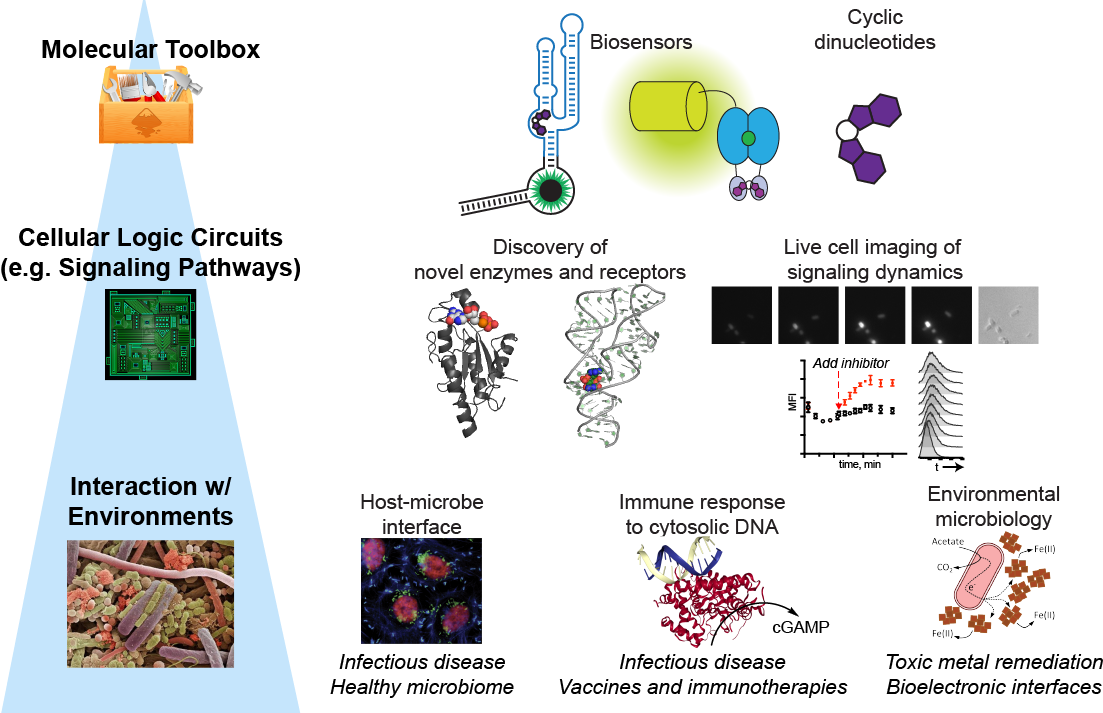
Molecular Imaging: RNA-based fluorescent biosensors
We are one of the first labs to develop fluorescent biosensors made of RNA for live cell imaging of enzyme activity. These sensors are designed by combining a riboswitch domain, which is an RNA that changes conformation upon binding a small molecule ligand, and a fluorophore-binding domain. Ligand selectivity is dictated by the riboswitch domain and can even be reprogrammed with single nucleotide changes.
Our lab has established allosteric stem design rules, novel designs, and optimization strategies for making RNA-based fluorescent biosensors from a variety of riboswitch folds. We have made biosensors with best-in-class sensitivity (sub-nanomolar limit of detection), selectivity (better than commercial monoclonal antibodies), and in vivo brightness (143% brighter than parent fluorophore-binding aptamer). We have applied these biosensors to visualize enzyme activity in both gram positive and gram negative bacteria, under aerobic and anaerobic conditions, via fluorescence microscopy and flow cytometry methods. We have 'watched' the chemical inhibition of an enzyme involved in biosynthesis of a quorum signaling molecule in live bacterial cells. We recently showed that the micronutrient zinc turns off a signaling enzyme that controls biofilm formation in E. coli, which helps to sensitize this common foodborne pathogen to antibiotics (Future of Biochemistry).
Biosensor Papers: JACS 2013a, JACS 2015, JACS 2016, NAR 2016, Cell Chem Biol 2016, Biochem 2018, Methods 2018
Other Riboswitch-Based Methods Papers: JACS 2013b, Chem Biol 2013, Anal Chem 2014, RNA Biol 2015
Reviews: Methods Enz 2015, Methods Mol Biol 2015, Methods Mol Biol 2017, Annu Rev Biochem 2017, Microbiol Spec 2018
Signaling: Cyclic dinucleotides in bacteria and mammalian cells
Cyclic dinucleotides are an emerging class of intracellular signaling molecules with diverse roles in bacteria and with newfound roles in innate immunity in mammalian cells. As second messengers, they are produced transiently in response to environmental stimuli and act inside the cell to change gene expression, physiology, and behavior. Given that three out of the four known cyclic dinucleotides were discovered in the past decade, much remains to be explored about their chemistry and biology. To study these chemical signals, our lab has developed fluorescent biosensors for each of the known cyclic dinucleotides. We also recently developed the first bioluminescent sensor for measuring cyclic dinucleotides in complex settings (Nat Chem Biol Research Highlight, JGI Science Highlight).
(i) Cyclic AMP-GMP (cAG) signaling: Using an in vitro biosensor assay, we discovered a cyclic AMP-GMP (cAG)-sensing riboswitch
class, which was highlighted by Science Signaling as a Signaling Breakthrough of the Year. These newfound cAG riboswitches were identified in Geobacter bacteria and predicted to control these bacteria's unique ability to relay electrons
outside the cell. By screening for enzyme activity using an in vivo biosensor assay,
we subsequently discovered a G. sulfurreducens GGDEF enzyme that makes cAG, which is the founding member of Hypr GGDEF enzymes.
This discovery is significant because for almost 30 years GGDEF enzymes were considered
synonymous with the classical cyclic di-GMP signaling network; our result provides
the first evidence that this enzyme class can make alternative cyclic dinucleotides
to cyclic di-GMP (Berkeley news story). We are continuing to identify molecular components of the cAG signaling pathway
and to understand its role in how bacteria like Geobacter sense and adapt to different kinds of surfaces.
(ii) Mammalian cGAMP signaling: The enzyme cyclic GMP-AMP synthase (cGAS) was discovered to be the innate immune
sensor for cytosolic DNA, which may result from microbial infection or other pathophysiological
conditions. Upon binding double-stranded DNA, cGAS produces the signaling molecule
cGAMP, which activates the receptor protein, stimulator of interferon genes (STING).
In collaboration with Russell Vance (UC Berkeley), we proved the chemical structure
of cGAMP had a noncanonical 2'-5' linkage by NMR. We recently developed a biosensor-based
platereader assay to quantitate cGAMP levels in DNA-stimulated mammalian cell lysates.
(iii) Other cyclic dinucleotide signaling: We have analyzed the activity of cyclic di-GMP and cyclic di-AMP signaling enzymes using in vivo biosensor assays. For example, we have demonstrated that archaeal enzymes produce cyclic di-AMP. This result completed the set of experimental evidence that cyclic dinucleotide signaling extends to all three domains of life.
cAG Signaling Papers: PNAS 2015, Cell Reports 2015, PNAS 2016
cGAMP Signaling Papers: Cell Reports 2013, Cell Chem Biol 2016
Other CDN Biosensor Papers: JACS 2013a, JACS 2015, NAR 2016, ACS Chem Bio 2018
