Nanopore Sequencing of DNA Damage
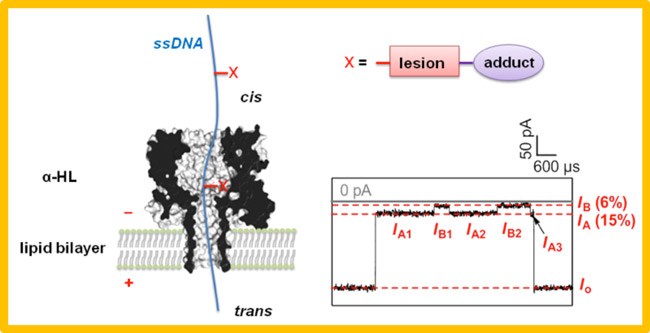
The protein ion channel ?-hemolysin provides the opportunity to study single molecules of DNA as they thread through the narrow constriction of the nanopore. Although this technology is of keen interest for single-molecule sequencing of DNA amplicons, we are focused on the investigation of native DNA that contains sites of modification or damage. For example, oxidative damage (OG) and base loss (AP sites) can be investigated in the channel by taking advantage of their unique chemistry in order to add bulky adducts. The adducts in turn give rise to modulations of the current while the DNA strand is electrophoretically driven through the pore. The ultimate goal is to determine the identity and sequence location of base modifications in single molecules of DNA which would be a tremendous breakthrough in the analysis of DNA damage.
Funding: NIH R01 GM093099
Collaborators: Henry White (University of Utah)--See photo of the Utah Nanopore Group

Additionally, we are investigating ways to make specific modifications to DNA strands to facilitate commercial efforts at DNA sequencing with the nanopore ion channel method.
Funding: NIH R01HG005095; DHS, DARPA
Collaborators: Geoff Barrall (Electronic Bio Sciences, San Diego); Eric Ervin and Anna Schibel (Electronic Bio Sciences, Salt Lake City)
