Base Switches for RNA Interference
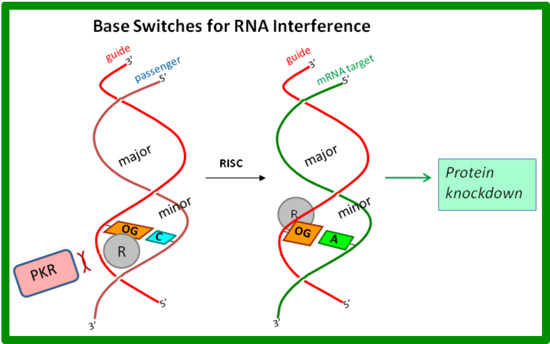
Selective nuclease digestion of messenger RNAs inside living cells via the short interfering RNA (siRNA)-triggered RNA interference pathway has become a mainstay in molecular biology to study gene function, and it holds promise for the development of new therapeutics. However, gaps in our understanding of the basic RNA interference mechanism and ability of siRNAs to interact with intracellular RNA-binding proteins, particularly those involved in the innate immune response, limit the application of this promising new technology. Our laboratory is developing new synthetic approaches to modified RNA oligonucleotides that carry out gene silencing with reduced undesirable off-target effects. Toward this end, we are developing modified bases capable to switching their steric blockades from the minor groove to the major groove (see figure). These modifications are placed in the antisense strand using a Watson-Crick-paired sense strand for delivery; the antisense strand is designed to target a complementary mRNA sequence via Hoogsteen base pairing in which a conformational change hides the steric blockade in the major groove.
Funding: NIH R01 GM080784
Collaborators: Peter A. Beal (UC-Davis)
